Example Burgers' equation
In this notebook we provide a simple example of the DeepMoD algorithm by applying it on the Burgers' equation.
We start by importing the required libraries and setting the plotting style:
# General imports
import numpy as np
import torch
import matplotlib.pylab as plt
# DeepMoD functions
from deepymod import DeepMoD
from deepymod.model.func_approx import NN
from deepymod.model.library import Library1D
from deepymod.model.constraint import LeastSquares
from deepymod.model.sparse_estimators import Threshold,PDEFIND
from deepymod.training import train
from deepymod.training.sparsity_scheduler import TrainTestPeriodic
from scipy.io import loadmat
# Settings for reproducibility
np.random.seed(42)
torch.manual_seed(0)
%load_ext autoreload
%autoreload 2
Next, we prepare the dataset.
data = np.load('data/burgers.npy', allow_pickle=True).item()
print('Shape of grid:', data['x'].shape)
Shape of grid: (256, 101)
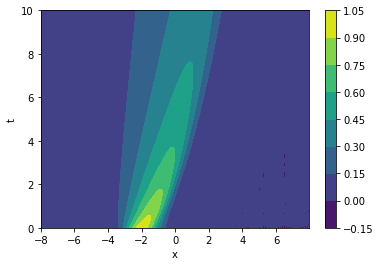
Let's plot it to get an idea of the data:
fig, ax = plt.subplots()
im = ax.contourf(data['x'], data['t'], np.real(data['u']))
ax.set_xlabel('x')
ax.set_ylabel('t')
fig.colorbar(mappable=im)
plt.show()
X = np.transpose((data['t'].flatten(), data['x'].flatten()))
y = np.real(data['u']).reshape((data['u'].size, 1))
print(X.shape, y.shape)
(25856, 2) (25856, 1)
As we can see, X has 2 dimensions, \{x, t\}, while y has only one, \{u\}. Always explicity set the shape (i.e. N\times 1, not N) or you'll get errors. This dataset is noiseless, so let's add 2.5\% noise:
noise_level = 0.025
y_noisy = y + noise_level * np.std(y) * np.random.randn(y[:,0].size, 1)
The dataset is also much larger than needed, so let's hussle it and pick out a 1000 samples:
number_of_samples = 2000
idx = np.random.permutation(y.shape[0])
X_train = torch.tensor(X[idx, :][:number_of_samples], dtype=torch.float32, requires_grad=True)
y_train = torch.tensor(y_noisy[idx, :][:number_of_samples], dtype=torch.float32)
print(X_train.shape, y_train.shape)
torch.Size([2000, 2]) torch.Size([2000, 1])
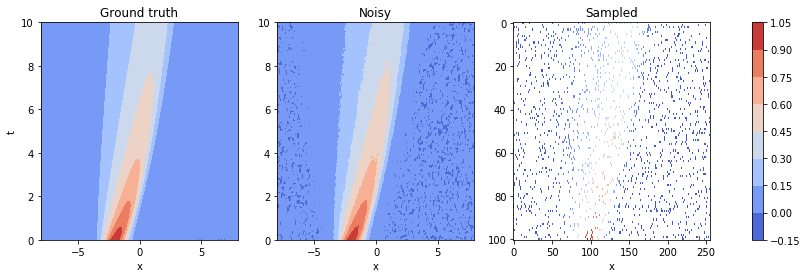
We now have a dataset which we can use. Let's plot, for a final time, the original dataset, the noisy set and the samples points:
fig, axes = plt.subplots(ncols=3, figsize=(15, 4))
im0 = axes[0].contourf(data['x'], data['t'], np.real(data['u']), cmap='coolwarm')
axes[0].set_xlabel('x')
axes[0].set_ylabel('t')
axes[0].set_title('Ground truth')
im1 = axes[1].contourf(data['x'], data['t'], y_noisy.reshape(data['x'].shape), cmap='coolwarm')
axes[1].set_xlabel('x')
axes[1].set_title('Noisy')
sampled = np.array([y_noisy[index, 0] if index in idx[:number_of_samples] else np.nan for index in np.arange(data['x'].size)])
sampled = np.rot90(sampled.reshape(data['x'].shape)) #array needs to be rotated because of imshow
im2 = axes[2].imshow(sampled, aspect='auto', cmap='coolwarm')
axes[2].set_xlabel('x')
axes[2].set_title('Sampled')
fig.colorbar(im1, ax=axes.ravel().tolist())
plt.show()
Configuring DeepMoD
Configuration of the function approximator: Here the first argument is the number of input and the last argument the number of output layers.
network = NN(2, [30, 30, 30, 30], 1)
Configuration of the library function: We select athe library with a 2D spatial input. Note that that the max differential order has been pre-determined here out of convinience. So, for poly_order 1 the library contains the following 12 terms: * [1, u_x, u_{xx}, u_{xxx}, u, u u_{x}, u u_{xx}, u u_{xxx}, u^2, u^2 u_{x}, u^2 u_{xx}, u^2 u_{xxx}]
library = Library1D(poly_order=2, diff_order=3)
Configuration of the sparsity estimator and sparsity scheduler used. In this case we use the most basic threshold-based Lasso estimator and a scheduler that asseses the validation loss after a given patience. If that value is smaller than 1e-5, the algorithm is converged.
estimator = Threshold(0.1)
sparsity_scheduler = TrainTestPeriodic(periodicity=50, patience=200, delta=1e-5)
Configuration of the sparsity estimator
constraint = LeastSquares()
# Configuration of the sparsity scheduler
Now we instantiate the model and select the optimizer
model = DeepMoD(network, library, estimator, constraint)
# Defining optimizer
optimizer = torch.optim.Adam(model.parameters(), betas=(0.99, 0.99), amsgrad=True, lr=1e-3)
Run DeepMoD
We can now run DeepMoD using all the options we have set and the training data: * The directory where the tensorboard file is written (log_dir) * The ratio of train/test set used (split) * The maximum number of iterations performed (max_iterations) * The absolute change in L1 norm considered converged (delta) * The amount of epochs over which the absolute change in L1 norm is calculated (patience)
train(model, X_train, y_train, optimizer,sparsity_scheduler, log_dir='runs/Burgers/', split=0.8, max_iterations=100000)
13350 MSE: 2.53e-05 Reg: 1.38e-05 L1: 1.45e+00 Algorithm converged. Writing model to disk.
Sparsity masks provide the active and non-active terms in the PDE:
model.sparsity_masks
[tensor([False, False, True, False, False, True, False, False, False, False,
False, False])]
estimatior_coeffs gives the magnitude of the active terms:
print(model.estimator_coeffs())
[array([[ 0. ],
[ 0. ],
[ 0.39227325],
[ 0. ],
[ 0. ],
[-1.001875 ],
[ 0. ],
[ 0. ],
[ 0. ],
[ 0. ],
[ 0. ],
[ 0. ]], dtype=float32)]
So the final terms that remain are the u_{xx} and u u_{x} resulting in the following Burgers equation (in normalized coefficients: u_t = 0.4 u_{xx} - u u_{x}.